As I have mentioned in my previous blog, in this blog I will be talking about LightGBM. However, this is not entirely about LightGBM (LGBM). Rather, this blog also includes other machine learning approaches. For the purpose of comparing results of SVM approach with non-SVM approaches, I borrowed the data obtained in a previous research we conducted last year, the results of which are published in this paper. We’ve had interesting results so I encourage you to please read the paper. This time, we used Random Forest (RF), XGBoost (XGB), and LightGBM (LGBM) to try to predict ligands of orphan GPCRs. I will not be delving into the details of each technique because there are already tons of information out there on the web that could explain them better. Instead, I will be discussing a comparison of these techniques being applied to the prediction of ligands of orphan GPCRs. So now, let’s get into it.
Summary of the study
Simply, we would like to know if we can accurately predict ligands of orphan targets. We only removed the training data for one particular target per model so we actually created 52 predictive models each corresponding to a virtual orphan target. Each model was used to screen all the compounds included in the original full model (no orphan targets) against each virtual orphan target. We then calculated the enrichment factor based on the top scoring 1% and also AUROC.
Training data
The training data utilized were, originally, from ChEMBL27 and are, specifically, for GPCR targets only. We have generated the positive and negative training data from previously created high activity (HA) CGBVS (SVM) predictive model for GPCRs. These training data were, basically, vectorized compound and protein descriptors generated using alvaDesc and multiple sequence alignment, respectively.
Predictive models
Fifty-two (52) predictive models were created corresponding to 52 GPCR targets for each of the 4 machine learning techniques (SVM, RF, XGB, LGBM) for a total of 208 predictive models. A predictive model from which the training data for a particular GPCR was omitted (virtual orphan GPCR) was used to screen approximately 280 thousand compounds (GPCR related) against the virtual orphan GPCR. The general workflow of the process is shown in Fig. 1.
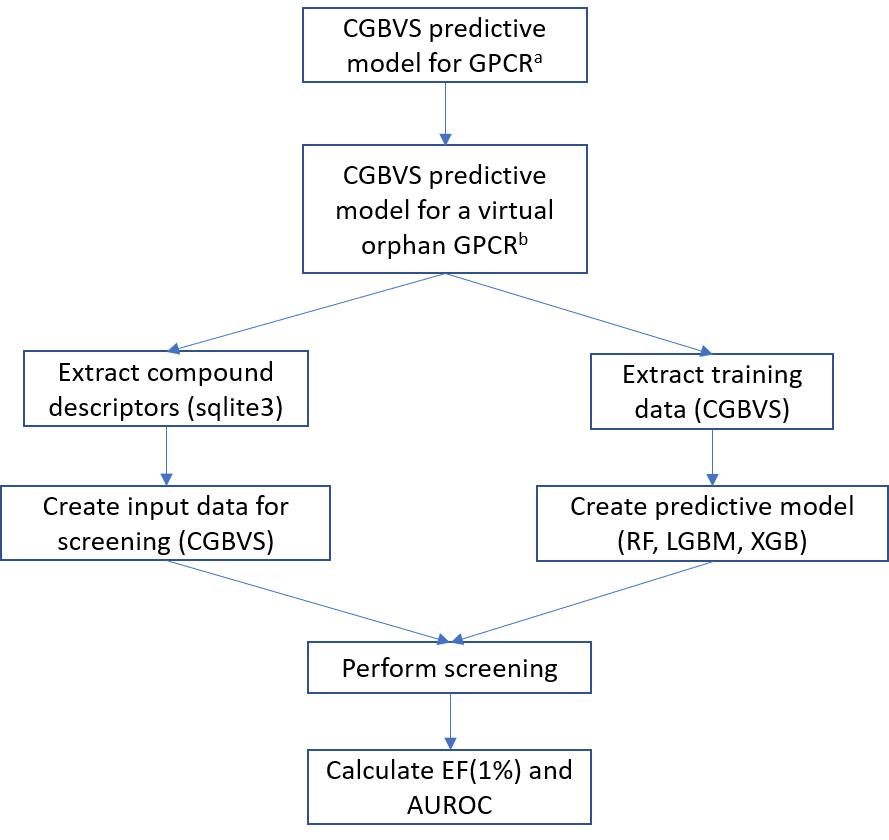
Figure 1. General workflow of the study. aGPCR model created using all available training data. bGPCR model created after omission of training data for a selected target protein.
Model creation was different with regard to SVM. SVM models were built using a CGBVS application (CzeekS) which was created using C++ programming language. The training data for the creation of RF, XGB, and LGBM models were derived from the SVM models themselves and the models were created using Python libraries. RF models were based on sklearn.ensemble’s RandomForestClassifier and on the other hand, XGB and LGBM models were based on the xgboost and lightgbm classifiers, respectively. Optimizations were performed using GPyOpt (RF) and Optuna (XGB, LGBM) and all work were done within an Anaconda environment. Graphs were created using R software.
Results
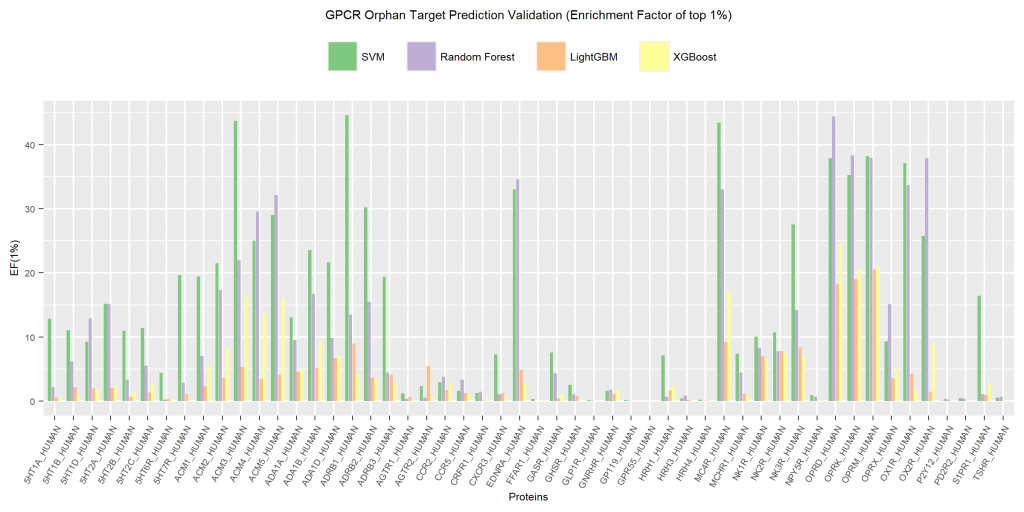
Calculated EF(1%) is shown in Fig. 2. The graph indicates that, in most cases, SVM models exhibited the best accuracy in predicting ligands of orphan targets, followed by RF with XGB and LGBM far behind.
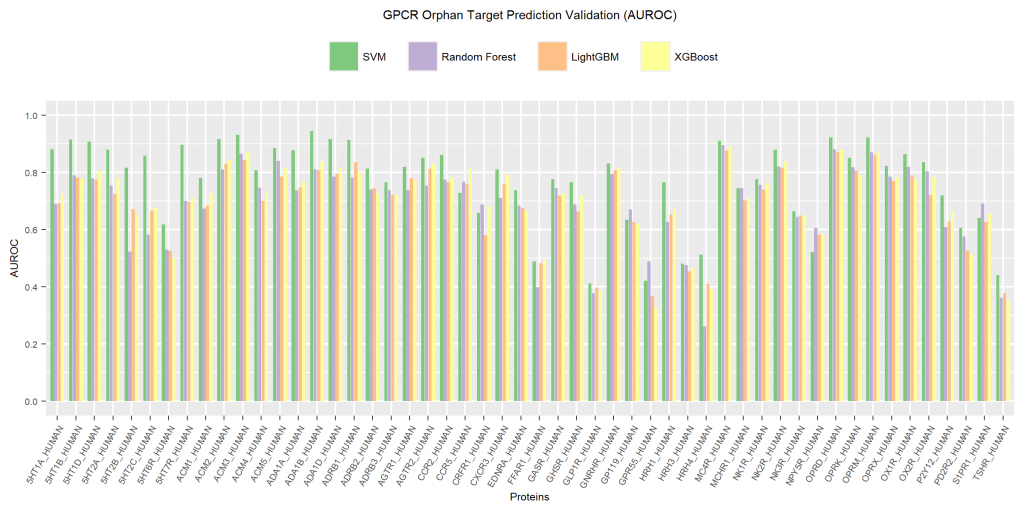
AUROC values also showed similar results with SVM exhibiting the best performance with maximum AUROC value around 0.95. Also in most cases, RF, XGB and LGBM values do not differ much. It probably has something to do with all of them being tree-based machine learning algorithms. It is also evident from the graph that targets belonging to a group with several related proteins showed higher AUROC values than those for singletons. This indicates that the existence or lack of training data for multiple related targets infuences ligand prediction accuracy for orphan targets.
I think it is safe to conclude that a tree based approach to create a model for the purpose of predicting ligands of orphan targets may not be the best course of action. Time consuming as it is, our original approach of using an SVM based model via CGBVS can be very promising for the same purpose. Maybe, another approach like Kernel Nearest Neighbor (KNN) method might also be promising.
Category: CGBVS/CzeekS, DRAGON/alvaDesc, Machine Learning